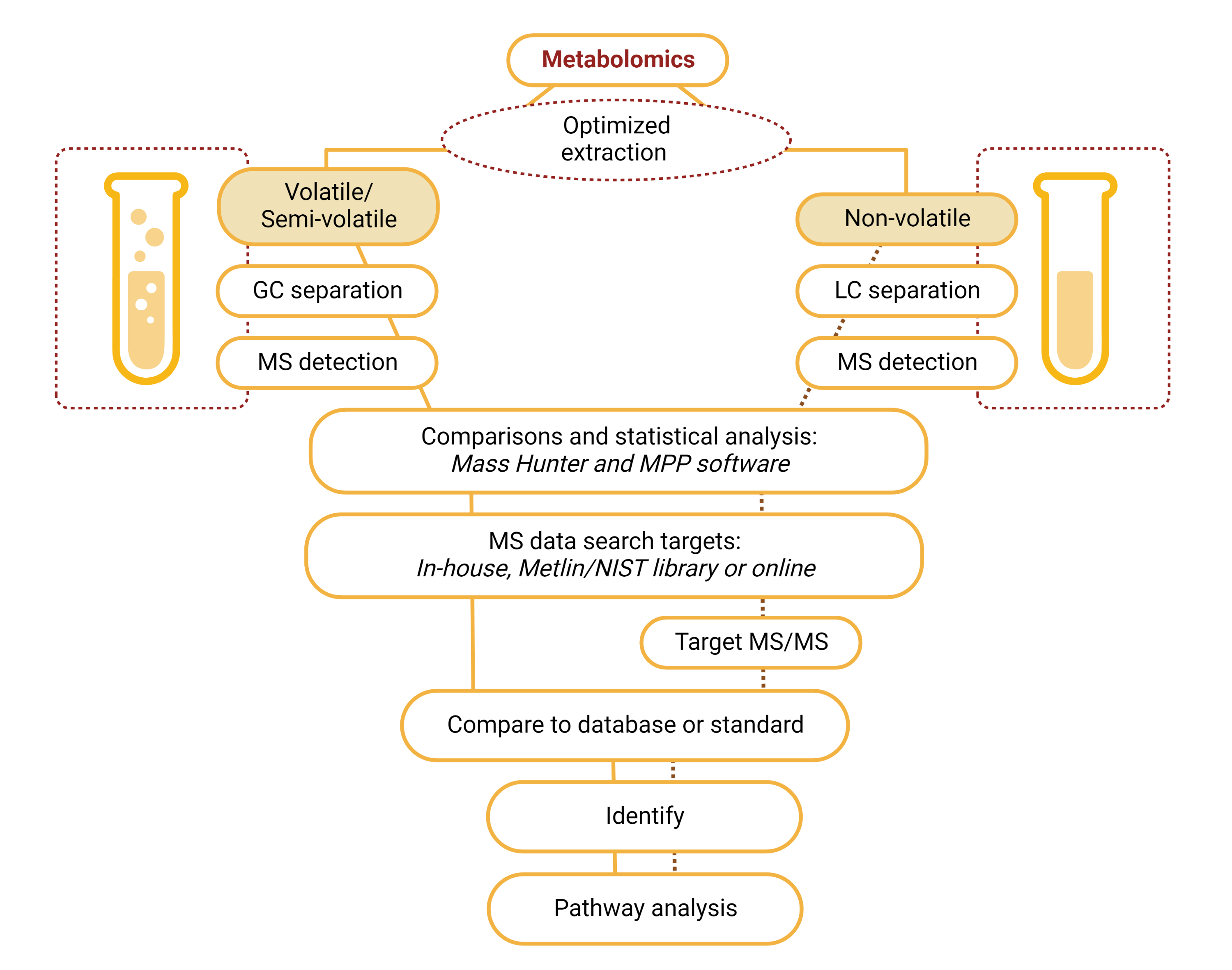
Metabolomics is the profiling of small molecules (<1500 Da) of a given sample at a specific time. Metabolites can be endogenous or exogenous, involved in primary or secondary metabolism. Untargeted metabolomics requires matrix and general target dependent sample extraction performed by user in advance. Data analysis is also specialized, MSF has a license for the Agilent Mass Profile Professional software there is also free software available. Access and training on MPP is available for a fee. Helpful references:
https://www.nature.com/articles/nrm.2016.25
https://www.embl.org/groups/metabolomics/protocols-used-for-lc-ms-analysis/
https://link.springer.com/book/10.1007/978-1-59745-244-1
Please consult MSF on these projects in advance.
Metabolomics Decision Tree