To label or not to label?
Qualitative proteomics can compare relative quantities of individual proteins in complex samples using a label-free approach or incorporate the use of protein or peptide labels.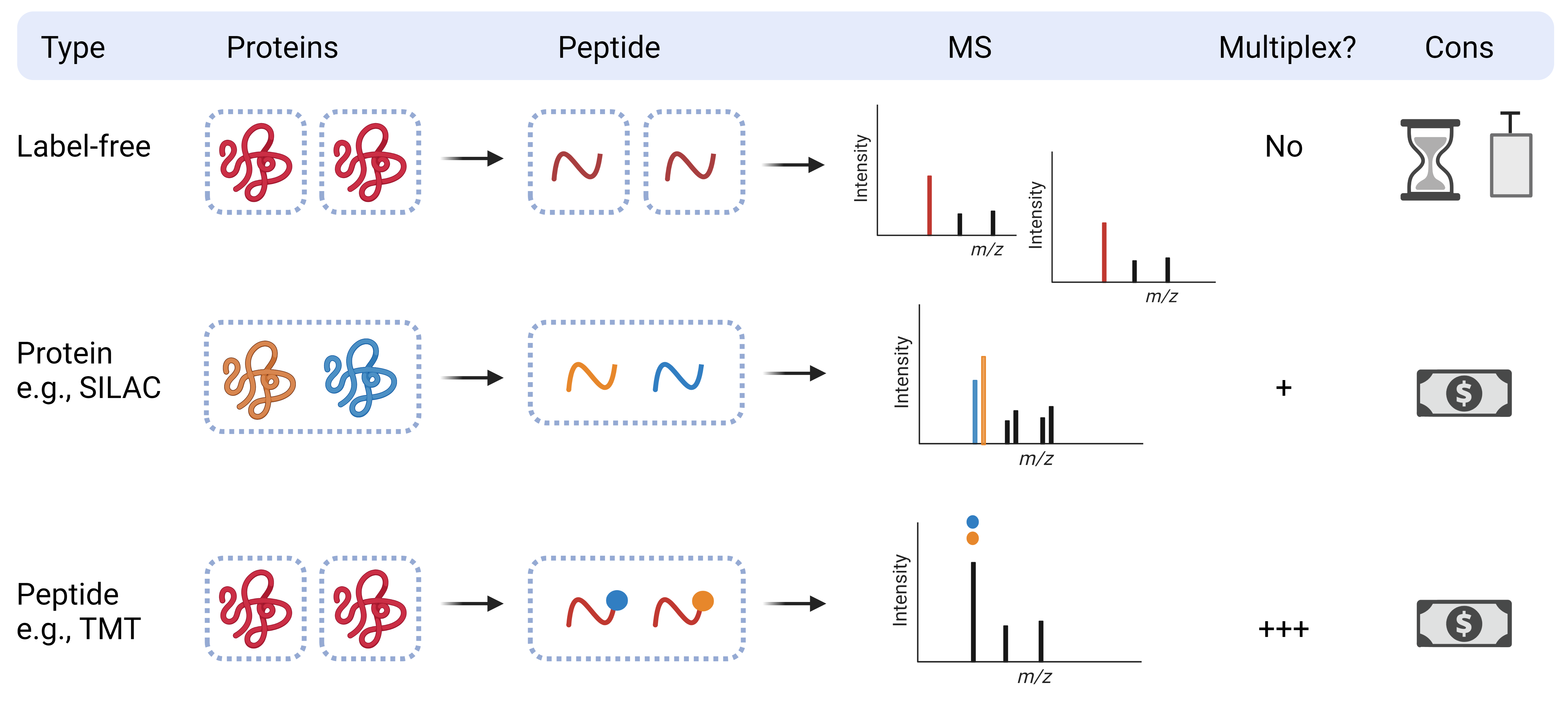
Discovery-based Quantitative MS methods
Quantitative Proteomics to Label or not to Label
Targeted Quantitative Proteomics MS Methods
MSF LFQ Service: Requirements and Deliverables
Label-free quantitation (LFQ): Normalization is completed based on overall sample intensity (i.e., total ion chromatogram), or an internal standard (spiked-in or “housekeeping” proteins).
Label-based quantitation: Proteins or peptides are labelled to allow for multiplexing and more precise relative quantitation between samples. Instrument time and technical variation can be reduced (vs. LFQ), but reagent costs are higher.