Project Requirements of Clients:
MSF clients must provide replicate samples of digested protein (i.e., peptides). Minimum four replicates per sample group is recommended, and a starting concentration of 25 micrograms per sample for complex proteomes. All peptides must be de-salted prior to injection using C18 spin columns, ZipTipTM, or similar. Recommended protocol.
Alternately, undigested protein samples can be submitted and associated fees will be applied for all additional processing completed by MSF staff (e.g., pre-digestion quantitation, in-solution digestion, desalting, etc.). Please refer to the MS Fees page for additional details.
Project Deliverables from MSF:
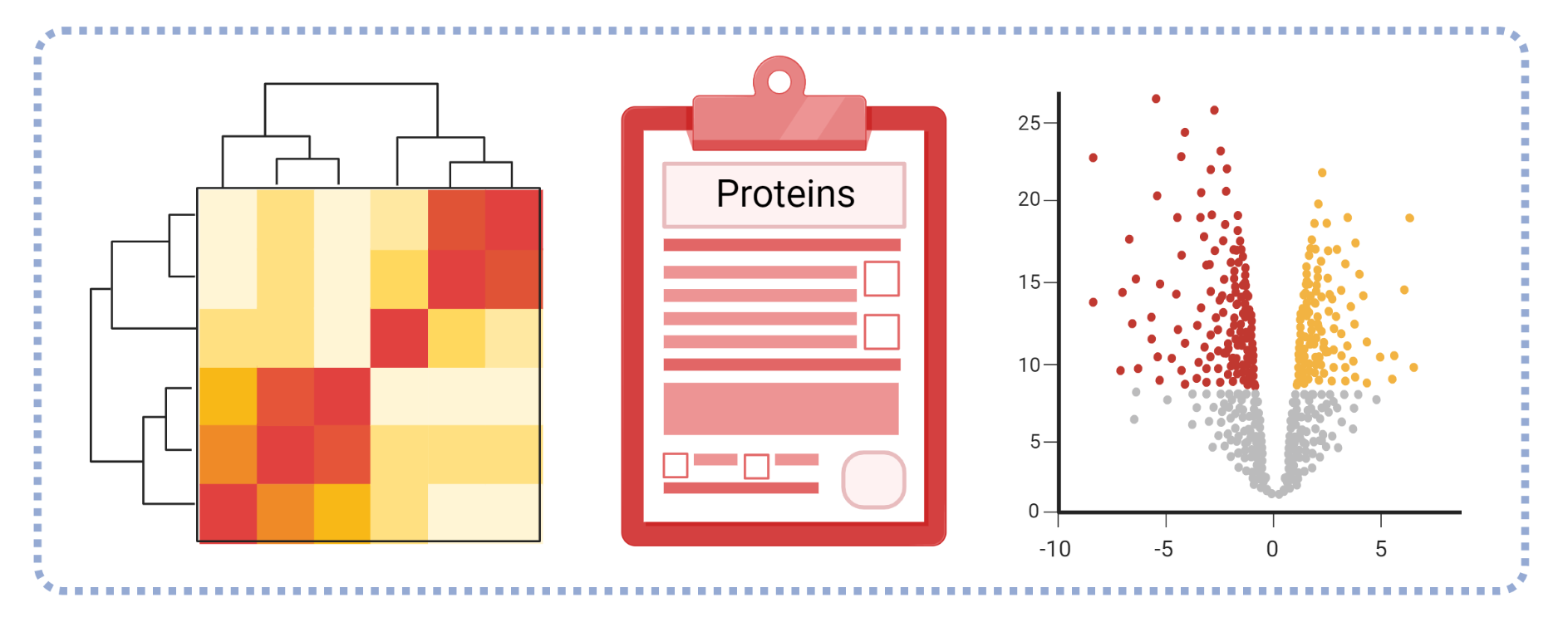
Upon completion, quantitative proteomics projects will include a set of raw data files and initial quantitative analysis with PEAKS Q software (Bioinformatics Solutions Inc., Waterloo, Ontario). Q exports include a volcano plot of significantly different proteins, protein profile heatmap, protein identifications and sample (or group) ratios, and a list of supporting peptides. Completed PEAKS projects can be viewed with a free downloadable viewer.
Add ons: Sample processing, peptide labelling, PEAKS software training, use and/or training with alternate analysis tools (e.g., MaxQuant and Perseus, Skyline).
Requirements and Deliverables may vary for other (i.e. non-LFQ) services. Please contact the MSF for more information.
PLEASE REVIEW https://www.uoguelph.ca/aac/important-considerations-each-stage-quantitative-proteomics
Discovery-based Quantitative MS methods
Quantitative Proteomics to Label or not to Label