AHL Newsletter, June 2014
For a pdf copy of AHL Newsletter, June 2014 click here
May 1, 2014, AHL User’s Guide and Fee Schedule
Effective May 1, 2014 we have updated our tests and fees. We have inactivated many of our species specific influenza tests for more “generic” labelling—whether avian, swine, equine, or canine, we’re looking for Influenza A virus.
We’ve added in many other new or modified tests over the past year: Bacterial culture/fecal culture equine, Brachyspira panel PCR, Hatchery reactor/excess mortality Salmonella, MLST molecular typing, Mycoplasma isolate prep for autogenous vaccine, Mycoplasma hyosynoviae and hyorhinis PCR, Potomac horse fever PCR, companion/other only zinc, copper and lead, Iodine in milk ICP-MS, Equid herpesvirus 1 PCR, PDCoV PCR, PEDV PCR, PEDV/TGEV/PRCV triplex PCR, Astrovirus PCR, and Bovine respiratory virus panel triplex PCR.
We have inactivated many older or less popular tests in favor of more suitable PCR tests.
Our complete test list is available in our May 1, 2014 AHL User’s Guide and Fee Schedule recently sent out to our clients. For a searchable test listing and pricing, our clients can visit us at our website at www.ahl.uoguelph.ca If you do not have client access for fees – please contact ahlinfo@uoguelph.ca , or call 519 824 4120 ext. 54320 and we will be more than happy to help you with this or anything else you might require.
Once again, we look forward to serving you!
Kudos!
Within the Animal Health Laboratory, we have many lab sections and must route samples appropriately. One client in particular, Mitchell Veterinary Services and their RVT Rhonda Kaufman, helps make this process seamless. Rhonda not only labels each specimen appropriately, and sends a legible history, she also splits the specimens according to the lab requirements.
Thank you Rhonda and Mitchell Vets for taking great care in submitting cases, your efforts are greatly appreciated!!
To freeze or not to freeze?
Josepha DeLay
Freezing and subsequent thawing has a significant negative impact on the gross and histologic appearance of tissues and organs. Tissue artefact caused by freezing can mask lesions and obscure the cause of death in postmortem examinations. Freezing artefact is especially problematic in cases with lesions involving brain, spinal cord, and gastrointestinal tract.
If an animal may be submitted for postmortem examination within 3 days of death / euthanasia, freezing should be avoided. Every effort should be made to deliver the body to the AHL as soon as possible after death, in order to preserve lesions and maximize the diagnostic value of postmortem examination. Refrigeration is preferable to freezing for storage of the body prior to delivery to the diagnostic laboratory. In situations where the interval between death and the postmortem exam will be prolonged (>3 days), freezing of the body is a viable option to prevent the equally compromising effects of autolysis on postmortem results.
AHL staff and pathologists are always available to offer advice regarding appropriate storage conditions for various clinical circumstances – please call us.
DSP Disease Surveillance Plan update
Grant Maxie
The AHL has been funded by the Ontario Ministry of Agriculture and Food - Ministry of Rural Affairs for the years 2013-2018 to develop and implement a “Disease Surveillance Plan”.
Vision:
Provide value to stakeholders through an effective and sustainable animal health network.
Objectives:
1. Develop an overall plan for the Ontario Animal Health Network (OAHN), and engage stakeholders.
2. Enhance Ontario animal disease surveillance.
3. Integrate Ontario animal disease surveillance with national surveillance.
4. Develop enhanced laboratory tools for early disease detection and surge capacity.
Progress to date includes:
- The formative stakeholder meeting was held at the Arboretum on the Guelph campus on September 10, 2013, with a theme of “Staying ahead of the curve” (the epidemic curve, that is!).
Presentations given at the meeting and feedback received from participants is posted at http://www.uoguelph.ca/omafra_partnership/en/partnershipprograms/DiseaseSurveillancePlanDSP.asp
- Appointed Dr. Maria Spinato as Deputy AHL Director to back up Dr. Grant Maxie, DSP project manager.
- Hired Andrew Vince, DVM, DVSc, Diplomate ACVP, to provide enhanced pathology input to client interactions.
- Conducted an environmental scan of client education resources.
- Hired Bruce McNab, DVM, PhD, as our part-time consulting epidemiologist.
- Began collaboration with a cross-Canada technical working group planning integration of selected provincial laboratory surveillance data into the Canadian Animal Health Surveillance Network (CAHSN).
- Hired 5 laboratory technicians to expand our technical resource and assist with development and implementation of enhanced testing, e.g., expanded molecular testing panels.
- Consulted with MAPAQ re RAIZO (Quebec Alert and Animal Health Information Network), which has a 20-year track record, and excellent results.
- Hired Melanie Barham, DVM, as Animal Health Network Coordinator to develop and implement expert networks in Ontario serving various sectors - swine, poultry, bovine, small ruminants, equine, camelids, cervids, fish, bees, companion animals.
- Continued discussions with the various Ontario veterinary organizations and commodity about how to best provide value to them through the DSP.
Antimicrobial susceptibility of mastitis pathogens isolated in the AHL mastitis section in 2012 and 2013
Durda Slavic, Beverly McEwen
There is a general lack of publicly available information regarding antimicrobial susceptibility of bacterial pathogens isolated from milk samples in Ontario. This certainly can be viewed as a disadvantage to practitioners who are using empirical therapy for mastitis cases. To provide our clients with some information regarding susceptibility patterns of common mastitis pathogens, the AHL laboratory information system was searched for years 2012 and 2013 and data are summarized in Tables 1A, 1B, 2A and 2B. The susceptibility results were available for over 2,700 isolates and data are shown as % resistant.
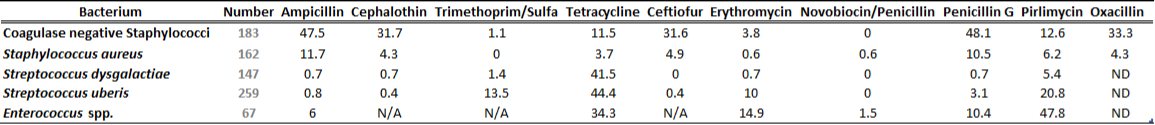
Data for gram-positive bacteria including CNS, S. aureus, Streptococcus dysgalactiae, Streptococcus uberis, and Enterococcus spp. are summarized in Tables 1A and 1B. Oxacillin susceptibility testing was done for coagulase-negative Staphylococcus spp. (CNS) and S. aureus only, whereas cephalosporins and trimethoprim/sulfa results are not applicable for Enterococcus spp. Enterococcus spp. must always be reported resistant to these drugs regardless of in vitro results. In general, CNS tend to be more resistant than S. aureus. Over 40% of isolates were resistant to penicillin, ampicillin, and oxacillin.
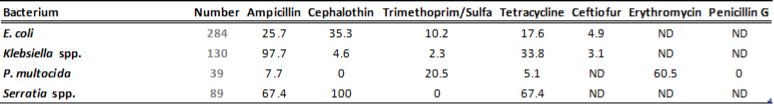
Data for gram-negative bacteria including Escherichia coli, Klebsiella spp., Serratia spp., and Pasteurella multocida are summarized in Tables 2A and 2B. Erythromycin and penicillin were tested only against P. multocida since they are not effective against other gram-negative bacteria specified below.
When extrapolating from data such as the ones shown below, one should exercise caution. It should be kept in mind that laboratory data in general are biased, being mostly associated with clinical cases and likely previous treatments. For that reason, the level of resistance in our data may be higher than in the average population. In addition, from the clinical perspective, our data are based on in vitro results. These results may or may not be reproduced in vivo. For example, both S. aureus and Serratia spp. can frequently be sensitive to some or all antimicrobials in vitro but will not respond to the treatment in vivo, particularly not in mastitis cases.
Table 1A. Susceptibility results for gram-positive bacteria isolated from milk samples in 2012. The results are expressed as % of resistant.
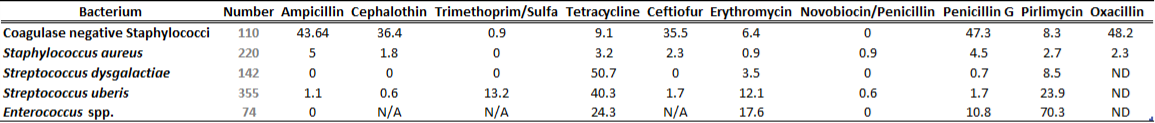
Table 1B. Susceptibility results for gram positive bacteria isolated from milk samples in 2013. The results are expressed as % of resistant.
ND=not done; N/A=not applicable
Table 2A. Susceptibility results for gram-negative bacteria isolated from milk samples in 2012. The results are expressed as % of resistant.
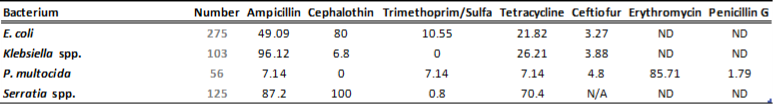
Table 2B. Susceptibility results for gram-negative bacteria isolated from milk samples in 2013. The results are expressed as % of resistant.
ND=not done; N/A=not applicable




